Karenina¶
Simulation and modeling tools for studying Anna Karenina effects in animal microbiomes This package aims to develop tools for modeling microbiome variability in disease. Initial versions focus on simulating microbiome change over time using simple Ornstein-Uhlenbeck (OU) models.
- Happy families are all alike; every unhappy family is unhappy in its own way.
- Leo Tolstoy
Links¶
Installation¶
Karenina is in active development and is available as a direct command-line program (karenina), or as a Qiime2 plugin (q2-karenina). Karenina is a python3.5+ program and requires scipy, pandas, matplotlib, and seaborn.
- Install ffmpeg (visualization dependency)
sudo apt-get install ffmpeg -y
- Install karenina
pip install git+https://github.com/zaneveld/karenina.git
Qiime2 Users¶
Qiime2 users will have the ability to use their Q2 Artifacts and receive output compatible with other Q2 plugins. q2-karenina requires both ffmpeg and karenina.
- Install q2-karenina
pip install git+https://github.com/zaneveld/q2-karenina.git
- Update Qiime2 cache to enable newly installed plugin.
qiime dev refresh-cache
Usage¶
| Script | Description |
|---|---|
| spatial_ornstein_uhlenbeck | OU Simulation Modeling |
| fit_timeseries | Fit PCoA Data to OU Models |
| karenina_visualization | Visualize PCoA Timeseries |
| qiime karenina spatial-ornstein-uhlenbeck | Q2 application of OU Modeling |
| qiime karenina fit-timeseries | Q2 application of PCoA fitting |
| qiime karenina visualization | Q2 application of 3D Visualization |
karenina¶
spatial_ornstein_uhlenbeck¶
>spatial_ornstein_uhlenbeck -h
Usage: spatial_ornstein_uhlenbeck -o ./simulation_results
This script simulates microbiome change over time using Ornstein-Uhlenbeck
(OU) models. These are similar to Brownian motion models, with the exception
that they include reversion to a mean. Output is a tab-delimited data table
and figures.
Options:
--version show program's version number and exit
-h, --help show this help message and exit
Required options:
-o OUTPUT, --output=OUTPUT
the output folder for the simulation results
Optional options:
--pert_file_path=PERT_FILE_PATH
file path to a perturbation file specifying parameters
for the simulation results
--treatment_names=TREATMENT_NAMES
Comma seperated list of treatment named
[default:control,destabilizing_treatment]
-n N_INDIVIDUALS, --n_individuals=N_INDIVIDUALS
Comma-separated number of individuals to simulate per
treatment.Note: This value must be enclosed in quotes.
Example: "35,35". [default: 35,35]
-t N_TIMEPOINTS, --n_timepoints=N_TIMEPOINTS
Number of timepoints to simulate. (One number, which
is the same for all treatments) [default: 10]
-p PERTURBATION_TIMEPOINT, --perturbation_timepoint=PERTURBATION_TIMEPOINT
Timepoint at which to apply a perturbation. Must be
less than --n_timepoints [default: 5]
-d PERTURBATION_DURATION, --perturbation_duration=PERTURBATION_DURATION
Duration that the perturbation lasts. [default: 100]
--interindividual_variation=INTERINDIVIDUAL_VARIATION
Starting variability between individuals. [default:
0.01]
--delta=DELTA Starting delta parameter for Brownian motion and
Ornstein-Uhlenbeck processes. A higher number
indicates more variability over time. [default: 0.25]
-l L, --L=L Starting lambda parameter for Ornstein-Uhlenbeck
processes. A higher number indicates a greater
tendancy to revert to the mean value. [default: 0.2]
--fixed_start_pos=FIXED_START_POS
Starting x,y,z position for all points, as comma
separated floating point values, e.g. 0.0,0.1,0.2. If
not supplied, starting positions will be randomized
based on the interindividual_variation parameter
[default: none]
-v, --verbose
allows for verbose output [default: False]
The spatial_ornstein_uhlenbeck function takes in simulation parameters and develops a distance matrix, PCoA ordination, and metadata file for use in other processes. This tutorial will explore the functionality of the script, and provide sample input data for fit_timeseries and karenina_visualization.
The following parameters are used to generate a simulation using Ornstein Uhlenbeck and Brownian motion models:
-o [Direct/filepath/to/output/dir/]
--n_timepoints 50
--perturbation_timepoint 15
--n_individuals 3,3
The following parameters were used as their default input:
--treatment_names control,destabilizing_treatment
--pert_file_path os.path.abspath(resource_filename('karenina.data','set_xyz_lambda_zero.tsv'))
--perturbation_duration 100
--interindividual_variation 0.01
--delta 0.25
--L 0.20
After installing karenina, the following command is used to execute the simulation from within the output directory.
spatial_ornstein_uhlenbeck -o ./ --n_timepoints 50 --perturbation_timepoint 15 --n_individuals 3,3
This generates the following files:
- ordination.txt : Simulation PCoA data
- metadata.tsv : Simulation metadata file
- log.txt : Simulation parameters
- euclidean.txt : Euclidean distance-matrix of Simulation data
- simulation_video.mp4 : Visualization of simulated data
These files can be viewed here:
https://github.com/zaneveld/karenina/tree/master/data/outputs/OU_simulation_50.3.3
fit_timeseries¶
>fit_timeseries -h
Usage: fit_timeseries -o ./simulation_results
This script fits microbiome change over time to Ornstein-Uhlenbeck (OU)
models.Demo fitting an OU model using default parameters.
Options:
--version show program's version number and exit
-h, --help show this help message and exit
Required options:
-o OUTPUT, --output=OUTPUT
the output folder for the simulation results
Optional options:
--fit_method=FIT_METHOD
Global optimization_method to use
[default:basinhopping]
--pcoa_qza=PCOA_QZA
The input PCoA qza file
--metadata=METADATA
The input metadata tsv file, if not defined, metadata
will be extracted from PCoA qza
--individual=INDIVIDUAL
The host subject ID column identifier, separate by
comma to combine TWO columns
--timepoint=TIMEPOINT
The timepoint column identifier
--treatment=TREATMENT
The treatment column identifier
-v, --verbose
allows for verbose output [default: False]
The fit_timeseries process allows users to fit PCoA and Metadata information to Ornstein Uhlenbeck models.
Warning
This script does not allow for optimization of single-point observations, where individuals are only measured or treated at one specific timepoint. If subjects are applied treatments at a single timepoint, it may be necessary to manually modify the metadata file to apply a boolean label to “treatment x applied” or not.
Note
The optimal number of measured timepoints is roughly 50 per subject, based on benchmarking.
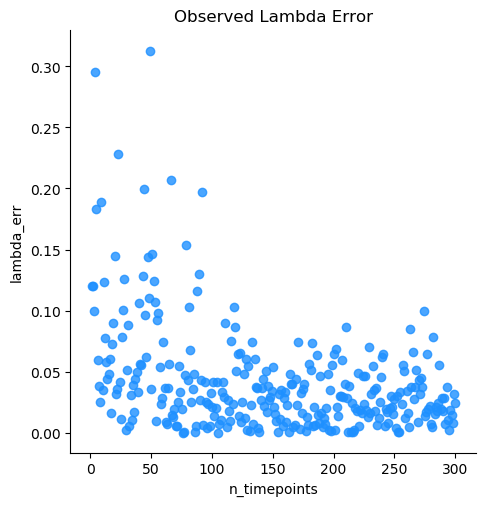
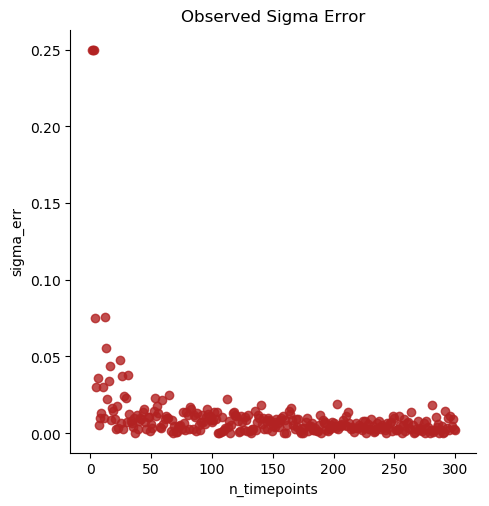

Using the data from the previously ran simulation (see spatial_ornstein_uhlenbeck), the fit_timeseries process will attempt to fit Ornstein Uhlenbeck models to the individuals and treatment cohorts of the input data.
The data being used for this tutorial can be found here:
https://github.com/zaneveld/karenina/blob/master/data/fit_timeseries/simulation.qza?raw=true
Note
fit_timeseries is currently configured to utilize basinhopping as its global optimizer, and more optimization methods will be added in the future.
fit_timeseries is currently configured to only accept Qiime2 formatted data, but will accept direct PCoA and metadata files in the future.
The following parameters are defined to fit the PCoA data to OU models:
-o [Direct/filepath/to/output/dir/]
--individual Subject
--timepoint Timepoint
--treatment Treatment
After installing karenina the following command may be used to generate OU model parameter output from within the same directory as simulation.qza. The output folder in this instance is /simulation_fit_ts.
fit_timeseries -o ./simulation_fit_ts/ \
--pcoa_qza ./simulation.qza \
--individual Subject \
--timepoint Timepoint \
--treatment Treatment
Note
fit_timeseries will create and optimize models for n_timepoints and n_individuals, and thus takes quite some time on larger datasets. Because this simulation data requires optimization of over 7000 entries, we have provided the data below.
With the treatment column identified, the program will generate two output files containing metadata for individuals and cohorts. Below is a sample of the cohort output generated. With input parameters being optimized to sigma/ delta: 0.25, lambda: 0.20, theta/ mu: 0.00 (from the OU simulation), the fit_timeseries process estimates the Ornstein Uhlenbeck model parameters.
This data can be found at the following link:
https://github.com/zaneveld/karenina/tree/master/data/outputs/fit_timeseries/tx_defined
Below is a sample of the cohort output generated, which estimates the control cohort to have parameters approaching the input. The perturbation set lambda to zero, and the estimation clearly displays the parameter approaching zero in the destabilizing_treatment cohort.
| Subject | pc | sigma | lambda | theta | nLogLik | aic |
|---|---|---|---|---|---|---|
| control_pc1 | pc1 | 0.25387 | 0.23143 | -0.0109 | -190.49 | -4.4992 |
| control_pc2 | pc2 | 0.25388 | 0.23144 | -0.0109 | -190.49 | -4.4992 |
| control_pc3 | pc3 | 0.25388 | 0.23144 | -0.0109 | -190.49 | -4.4992 |
| destabilizing_treatment_pc1 | pc1 | 0.24395 | 0.01951 | -0.1203 | -201.98 | -4.6163 |
| destabilizing_treatment_pc2 | pc2 | 0.24395 | 0.01951 | -0.1203 | -201.98 | -4.6163 |
| destabilizing_treatment_pc3 | pc3 | 0.24395 | 0.01951 | -0.1203 | -201.98 | -4.6163 |
Because the simulation data is large, and thus takes some time to process, we can alternatively fit the timeseries from Moving Pictures. The weighted_unifrac_pcoa_results.qza file will be used.
The PCoA data can be found here:
Because the Moving Pictures data contains single-point observations for their treatments, being that their treatments are applied at timepoint-0 only, fit_timeseries will fit OU models to only individuals if we do not define a treatment identifier.
After installing karenina the following command can be ran from the same directory as weighted_unifrac_pcoa_results.qza.
Note
The individual column combines the columns Subject and BodySite, this is entered as a comma-separated list.
Note
The treatments are single-point observations, so fit_timeseries will fit to only individuals if treatment-col is None.
fit_timeseries \
-o ./moving_pictures_fit_ts/
--pcoa_qza ./weighted_unifrac_pcoa_results.qza
--individual Subject,BodySite
--timepoint DaysSinceExperimentStart
The output data from this execution can be found here:
https://github.com/zaneveld/karenina/blob/master/data/outputs/fit_timeseries/tx_undefined
| Subject_BodySite | pc | sigma | lambda | theta | nLogLik | aic |
|---|---|---|---|---|---|---|
| subject-1_gut | pc1 | 0.41463 | 0.00360 | -0.5421 | -30.398 | 1.17122 |
| subject-1_gut | pc2 | -0.4051 | 0.03268 | 0.02503 | -30.398 | 1.17122 |
| subject-1_gut | pc3 | 0.42444 | 0.04177 | -0.0019 | -30.398 | 1.17122 |
| subject-1_left-palm | pc1 | -0.4043 | 0.02676 | 0.18309 | -30.398 | 1.17122 |
| subject-1_left-palm | pc2 | -0.4021 | -0.0245 | -0.2053 | -30.398 | 1.17122 |
| subject-1_left-palm | pc3 | 0.39130 | -0.0246 | 0.28571 | -30.398 | 1.17122 |
| subject-1_right-palm | pc1 | -0.4045 | 0.03113 | 0.15796 | -30.398 | 1.17122 |
| subject-1_right-palm | pc2 | 0.41556 | 0.00018 | -2.5981 | -30.398 | 1.17122 |
| subject-1_right-palm | pc3 | 0.41527 | -0.2349 | 0.13882 | -30.398 | 1.17122 |
| subject-1_tongue | pc1 | -0.8731 | 0.04062 | 0.27590 | -27.550 | 3.36793 |
| subject-1_tongue | pc2 | -0.8970 | 0.00520 | 0.29044 | -24.257 | 3.62250 |
| subject-1_tongue | pc3 | -0.8792 | 0.03174 | -0.0404 | -26.767 | 3.42560 |
| subject-2_gut | pc1 | 0.40325 | -0.0210 | -0.5796 | -30.398 | 1.17122 |
| subject-2_gut | pc2 | -0.4024 | 0.00664 | 0.01683 | -30.398 | 1.17122 |
| subject-2_gut | pc3 | -0.4132 | 0.01286 | 0.00126 | -30.398 | 1.17122 |
| subject-2_left-palm | pc1 | 0.39737 | 0.01596 | 0.18911 | -30.398 | 1.17122 |
| subject-2_left-palm | pc2 | 0.41552 | 0.03488 | 0.04242 | -30.398 | 1.17122 |
| subject-2_left-palm | pc3 | -0.4025 | -0.0174 | 0.09704 | -30.398 | 1.17122 |
| subject-2_right-palm | pc1 | -0.4023 | 0.00992 | 0.33786 | -30.398 | 1.17122 |
| subject-2_right-palm | pc2 | -0.4132 | 0.04034 | -0.0563 | -30.398 | 1.17122 |
| subject-2_right-palm | pc3 | 0.41496 | 0.00458 | -0.0590 | -30.398 | 1.17122 |
| subject-2_tongue | pc1 | -0.4038 | 0.46210 | 0.29795 | -30.398 | 1.17122 |
| subject-2_tongue | pc2 | -0.4145 | 0.33915 | 0.21338 | -30.398 | 1.17122 |
| subject-2_tongue | pc3 | 0.41374 | -0.1617 | 0.07803 | -30.398 | 1.17122 |
karenina_visualization¶
Warning
This function is currently experiencing some bugs, and is not recommended for use at this time. Visualizations can be generated through execution of spatial_ornstein_uhlenbeck, and is currently being addressed.
>karenina_visualization -h
Usage: karenina_visualization -o ./simulation_results
This script fits microbiome change over time to Ornstein-Uhlenbeck (OU)
models.Demo fitting an OU model using default parameters.
Options:
--version show program's version number and exit
-h, --help show this help message and exit
Required options:
-o OUTPUT, --output=OUTPUT
the output folder for the simulation results
Optional options:
--pcoa_qza=PCOA_QZA
The input PCoA qza file
--metadata=METADATA
The input metadata tsv file, if not defined, metadata
will be extracted from PCoA qza
--individual=INDIVIDUAL
The host subject ID column identifier, separate by
comma to combine TWO columns
--timepoint=TIMEPOINT
The timepoint column identifier
--treatment=TREATMENT
The treatment column identifier
-v, --verbose
allows for verbose output [default: False]
The karenina_visualization function allows users to visualize their PCoA results as an mp4 movie.
Using the data from the previously ran simulation (see spatial_ornstein_uhlenbeck), the karenina_visualization program will visualize the PCoA Results, and return an mp4 movie called simulation_video.mp4.
The data being used for this tutorial can be found here:
https://github.com/zaneveld/karenina/blob/master/data/fit_timeseries/simulation.qza?raw=true
The following parameters are defined to visualize the PCoA data:
-o [Direct/filepath/to/output/dir/]
--individual Subject
--timepoint Timepoint
--treatment Treatment
After installing karenina the following command may be used to generate a 3d visualization video from within the same directory as simulation.qza. The output folder in this instance is /simulation_vis.
karenina_visualization -o ./simulation_vis/ \
--pcoa_qza ./simulation.qza \
--individual Subject \
--timepoint Timepoint \
--treatment Treatment
The output video can be viewed below, and the file can be found here:
https://github.com/zaneveld/karenina/tree/master/data/outputs/visualization
q2-karenina¶
>qiime karenina --help
Usage: qiime karenina [OPTIONS] COMMAND [ARGS]...
Description: This script simulates microbiome change over time using
Ornstein-Uhlenbeck (OU) models. These are similar to Brownian motion
models, with the exception that they include reversion to a mean. Output
is a tab-delimited data table and figures.
Plugin website: https://github.com/zaneveld/karenina
Getting user support: Please post to the QIIME 2 forum for help with this
plugin: https://forum.qiime2.org
Options:
--version Show the version and exit.
--citations Show citations and exit.
--help Show this message and exit.
Commands:
fit-timeseries Fit OU Models to PCoA Ordination output
spatial-ornstein-uhlenbeck Spatial Ornstein Uhlenbeck microbial community
simulation
visualization Generates 3D animations of PCoA Timeseries
qiime karenina spatial-ornstein-uhlenbeck¶
Warning
This method is currently under construction and will be available for use in Qiime2 in the near future. to implement this method, please reference karenina spatial_ornstein_uhlenbeck.
>qiime karenina spatial-ornstein-uhlenbeck --help
Usage: qiime karenina spatial-ornstein-uhlenbeck [OPTIONS]
This method simulates microbial behavior over time usingOrnstein Uhlenbeck
models. This are similar to Brownian Motionwith the exception that they
include reversion to a mean.
Options:
--p-perturbation-fp TEXT
filepath for perturbation parameters for
simulation results [required]
--p-treatment-names TEXT
['control,destabilizing_treatment'] Names
for simulation treatments [required]
--p-n-individuals TEXT
['35,35'] Number of individuals per
treatment [required]
--p-n-timepoints INTEGER
['10'] Number of simulation timepoints
[required]
--p-perturbation-timepoint INTEGER
['5'] Timepoint at which to apply treatment
(<n_timepoints) [required]
--p-perturbation-duration INTEGER
['100'] Duration of perturbation.
[required]
--p-interindividual-variation FLOAT
['0.01']Starting variability between
individuals [required]
--p-delta FLOAT
['0.25'] Starting Delta parameter for
Brownian Motion/ OU models. Higher values
indicate greater variability over time
[required]
--p-lam FLOAT
['0.20'] Starting Lambda value for OU
process. Higher values indicate a greater
tendancy to revert to the mean value.
[required]
--p-fixed-start-pos TEXT
Starting x,y,z position for each point. If
not defined, starting positions will be
randomized based on
interindividual_variation; type: string, eg:
['0.0,0.1,0.2']. [required]
--o-ordination ARTIFACT PATH PCoAResults
Sample PCoA file containing simulation data
[required if not passing --output-dir]
--o-distance-matrix ARTIFACT PATH DistanceMatrix
Sample Distance Matrix containing simulation
data [required if not passing --output-dir]
--output-dir DIRECTORY
Output unspecified results to a directory
--cmd-config PATH
Use config file for command options
--verbose
Display verbose output to stdout and/or
stderr during execution of this action.
[default: False]
--quiet
Silence output if execution is successful
(silence is golden). [default: False]
--citations
Show citations and exit.
--help
Show this message and exit.
Note
The tutorial for this Q2 Method is being created and will be available in the future.
qiime karenina fit-timeseries¶
Note
This visualization currently does not accept Qiime2 artifacts, and is currently being addressed. Please note that the input is the filepath to the PCoA Results qza file.
>qiime karenina fit-timeseries --help
Usage: qiime karenina fit-timeseries [OPTIONS]
This visualizer generates OU model parameters for PCoA outputdata, for
each individual and each defined treatment cohort.
Options:
--p-pcoa TEXT
filepath to PCoA results [required]
--p-metadata TEXT
filepath to Sample metadata [required]
--p-method [basinhopping]
global optimization method [required]
--p-individual-col TEXT
individual column identifier [required]
--p-timepoint-col TEXT
timepoint column identifier [required]
--p-treatment-col TEXT
treatment column identifier [required]
--o-visualization VISUALIZATION PATH
[required if not passing --output-dir]
--output-dir DIRECTORY
Output unspecified results to a directory
--cmd-config PATH
Use config file for command options
--verbose
Display verbose output to stdout and/or
stderr during execution of this action.
[default: False]
--quiet
Silence output if execution is successful
(silence is golden). [default: False]
--citations
Show citations and exit.
--help
Show this message and exit.
The visualization allows for the use of Qiime 2 to fit Ornstein Uhlenbeck models to PCoA data. Utilizing previous simulation data which can be found at the following link, we will walk through the use of this plugin.
Warning
This script does not allow for optimization of single-point observations, where individuals are only measured or treated at one specific timepoint. If subjects are applied treatments at a single timepoint, it may be necessary to manually modify the metadata file to apply a boolean label to “treatment x applied” or not.
Note
The optimal number of measured timepoints is roughly 50 per subject, based on benchmarking.



The data being used for this tutorial can be found here:
https://github.com/zaneveld/karenina/blob/master/data/fit_timeseries/simulation.qza?raw=true
The following parameters are defined to fit the PCoA data to OU models:
Notice the metadata is within the qza file, so the metadata parameter is defined as None
--p-pcoa [Direct/filepath/to/pcoaResults.qza]
--p-metadata None
Note
The only optimization method employed at this time is basinhopping
--p-method basinhopping
Within the metadata file, we see that the column identifying individuals, timepoints, and treatment are Subject, Timepoint, Treatment.
We define the following parameters to match these identifiers
--p-individual-col Subject
--p-timepoint-col Timepoint
--p-treatment-col Treatment
The output directory is defined as such:
--output-dir /home/qiime2/simulation_ou_fit_ts/
After installing q2-karenina the following command may be used to generate OU model parameter output from within the same directory as simulation.qza.
qiime karenina fit-timeseries \
--p-pcoa ./simulation.qza \
--p-metadata None \
--p-method basinhopping \
--p-individual-col Subject \
--p-timepoint-col Timepoint \
--p-treatment-col Treatment \
--output-dir /home/qiime2/simulation_ou_fit_ts/
A successful visualization provides the following output:
Saved Visualization to: /home/qiime2/simulation_ou_fit_ts/visualization.qzv
Within the visualization.qzv, we have two output data files which contain our modeled timeseries results. With input parameters being optimized to sigma/ delta: 0.25, lambda: 0.20, theta/ mu: 0.00 (from the OU simulation), the fit_timeseries modeled individuals and cohorts, which can be found here:
https://github.com/zaneveld/q2-karenina/blob/master/data/simulation_ou_fit_ts
Below is a sample of the cohort output generated, which estimates the control cohort to have parameters approaching the input. The perturbation set lambda to zero, and the estimation clearly displays the parameter approaching zero in the destabilizing_treatment cohort.
| Subject | pc | sigma | lambda | theta | nLogLik | aic |
|---|---|---|---|---|---|---|
| control_pc1 | pc1 | 0.25387 | 0.23143 | -0.0109 | -190.49 | -4.4992 |
| control_pc2 | pc2 | 0.25388 | 0.23144 | -0.0109 | -190.49 | -4.4992 |
| control_pc3 | pc3 | 0.25388 | 0.23144 | -0.0109 | -190.49 | -4.4992 |
| destabilizing_treatment_pc1 | pc1 | 0.24395 | 0.01951 | -0.1203 | -201.98 | -4.6163 |
| destabilizing_treatment_pc2 | pc2 | 0.24395 | 0.01951 | -0.1203 | -201.98 | -4.6163 |
| destabilizing_treatment_pc3 | pc3 | 0.24395 | 0.01951 | -0.1203 | -201.98 | -4.6163 |
Because the simulation data is large, and thus takes some time to process, we can alternatively fit the timeseries from Moving Pictures. The weighted_unifract_pcoa_results.qza file will be used.
The PCoA data can be found here:
Because the Moving Pictures data contains single-point observations for their treatments, being that their treatments are applied at timepoint-0 only, fit_timeseries will fit OU models to only individuals if we do not define a treatment identifier.
After installing q2-karenina the following command can be ran from the same directory as weighted_unifrac_pcoa_results.qza.
Note
The individual column combines the columns Subject and BodySite, this is entered as a comma-separated list.
Note
The treatments are single-point observations, so fit_timeseries will fit to only individuals if treatment-col is None.
qiime karenina fit-timeseries \
--p-pcoa ./weighted_unifrac_pcoa_results.qza \
--p-metadata None \
--p-method basinhopping \
--p-individual-col Subject,BodySite \
--p-timepoint-col DaysSinceExperimentStart \
--p-treatment-col None \
--output-dir ./moving_pictures_fit_ts/
A successful visualization provides the following output:
Saved Visualization to: ./moving_pictures_fit_ts/visualization.qzv
The output data from this execution can be found here:
https://github.com/zaneveld/q2-karenina/blob/master/data/moving_pictures_fit_ts
| Subject_BodySite | pc | sigma | lambda | theta | nLogLik | aic |
|---|---|---|---|---|---|---|
| subject-1_gut | pc1 | 0.41463 | 0.00360 | -0.5421 | -30.398 | 1.17122 |
| subject-1_gut | pc2 | -0.4051 | 0.03268 | 0.02503 | -30.398 | 1.17122 |
| subject-1_gut | pc3 | 0.42444 | 0.04177 | -0.0019 | -30.398 | 1.17122 |
| subject-1_left-palm | pc1 | -0.4043 | 0.02676 | 0.18309 | -30.398 | 1.17122 |
| subject-1_left-palm | pc2 | -0.4021 | -0.0245 | -0.2053 | -30.398 | 1.17122 |
| subject-1_left-palm | pc3 | 0.39130 | -0.0246 | 0.28571 | -30.398 | 1.17122 |
| subject-1_right-palm | pc1 | -0.4045 | 0.03113 | 0.15796 | -30.398 | 1.17122 |
| subject-1_right-palm | pc2 | 0.41556 | 0.00018 | -2.5981 | -30.398 | 1.17122 |
| subject-1_right-palm | pc3 | 0.41527 | -0.2349 | 0.13882 | -30.398 | 1.17122 |
| subject-1_tongue | pc1 | -0.8731 | 0.04062 | 0.27590 | -27.550 | 3.36793 |
| subject-1_tongue | pc2 | -0.8970 | 0.00520 | 0.29044 | -24.257 | 3.62250 |
| subject-1_tongue | pc3 | -0.8792 | 0.03174 | -0.0404 | -26.767 | 3.42560 |
| subject-2_gut | pc1 | 0.40325 | -0.0210 | -0.5796 | -30.398 | 1.17122 |
| subject-2_gut | pc2 | -0.4024 | 0.00664 | 0.01683 | -30.398 | 1.17122 |
| subject-2_gut | pc3 | -0.4132 | 0.01286 | 0.00126 | -30.398 | 1.17122 |
| subject-2_left-palm | pc1 | 0.39737 | 0.01596 | 0.18911 | -30.398 | 1.17122 |
| subject-2_left-palm | pc2 | 0.41552 | 0.03488 | 0.04242 | -30.398 | 1.17122 |
| subject-2_left-palm | pc3 | -0.4025 | -0.0174 | 0.09704 | -30.398 | 1.17122 |
| subject-2_right-palm | pc1 | -0.4023 | 0.00992 | 0.33786 | -30.398 | 1.17122 |
| subject-2_right-palm | pc2 | -0.4132 | 0.04034 | -0.0563 | -30.398 | 1.17122 |
| subject-2_right-palm | pc3 | 0.41496 | 0.00458 | -0.0590 | -30.398 | 1.17122 |
| subject-2_tongue | pc1 | -0.4038 | 0.46210 | 0.29795 | -30.398 | 1.17122 |
| subject-2_tongue | pc2 | -0.4145 | 0.33915 | 0.21338 | -30.398 | 1.17122 |
| subject-2_tongue | pc3 | 0.41374 | -0.1617 | 0.07803 | -30.398 | 1.17122 |
qiime karenina visualization¶
Warning
This visualization is currently under construction and will be available for use in Qiime2 in the near future. to implement this method, please reference karenina_visualization.
>qiime karenina visualization --help
Usage: qiime karenina visualization [OPTIONS]
This visualizer generates 3D animations of PCoA Timeseries.
Options:
--p-pcoa TEXT
filepath to PCoA results [required]
--p-metadata TEXT
filepath to Sample metadata [required]
--p-individual-col TEXT
individual column identifier [required]
--p-timepoint-col TEXT
timepoint column identifier [required]
--p-treatment-col TEXT
treatment column identifier [required]
--o-visualization VISUALIZATION PATH
[required if not passing --output-dir]
--output-dir DIRECTORY
Output unspecified results to a directory
--cmd-config PATH
Use config file for command options
--verbose
Display verbose output to stdout and/or
stderr during execution of this action.
[default: False]
--quiet
Silence output if execution is successful
(silence is golden). [default: False]
--citations
Show citations and exit.
--help
Show this message and exit.
Note
The tutorial for this Q2 Visualization is being created and will be available in the future.
spatial_ornstein_uhlenbeck.py¶
-
karenina.spatial_ornstein_uhlenbeck.check_perturbation_timepoint(perturbation_timepoint, n_timepoints)¶ Raise ValueError if perturbation_timepoint is < 0 or >n_timepoints
Parameters: - perturbation_timepoint – defined timepoint for perturbation application
- n_timepoints – number of timepoints
-
karenina.spatial_ornstein_uhlenbeck.ensure_exists(output_dir)¶ Ensure that output_dir exists
Parameters: output_dir – path to output directory
-
karenina.spatial_ornstein_uhlenbeck.parse_perturbation_file(pert_file_path, perturbation_timepoint, perturbation_duration)¶ Return a list of perturbations infile – a .tsv file describing one perturbation per line assume input file is correctly formatted (no warnings if not)
NOTE: each pertubation should be in the format:
set_xyz_lambda_low = {“start”:opts.perturbation_timepoint, “end”:opts.perturbation_timepoint + opts.perturbation_duration, “params”:{“lambda”:0.005}, “update_mode”:”replace”, “axes”:[“x”,”y”,”z”]}
Parameters: - pert_file_path – perturbation file path
- perturbation_timepoint – timepoint to apply perturbation
- perturbation_duration – duration of perturbation
Returns: perturbation list parsed from pert file contents
-
karenina.spatial_ornstein_uhlenbeck.write_options_to_log(log, opts)¶ Writes user’s input options to log file
Parameters: - log – log filename
- opts – options
fit_timeseries.py¶
-
karenina.fit_timeseries.aic(n_param, nLogLik)¶ calculates AIC with 2 * n_parameters - 2 * LN(-1 * nLogLik)
Parameters: - n_param – number of parameters
- nLogLik – negative log likelihood
Returns: aic score
-
karenina.fit_timeseries.fit_cohorts(input, ind, tp, tx, method, verbose=False)¶ Completes the same operation as fit_input, for cohorts. Fit and minimize the timeseries for each subject and cohort defined.
Parameters: - input – dataframe: [#SampleID,individual,timepoint,treatment,pc1,pc2,pc3]
- ind – subject column identifier
- tp – timepoint column identifier
- tx – treatment column identifier
- method – “basinhopping” if not defined in opts
Returns: dataframe: [ind,”pc”,”sigma”,”lambda”,”theta”,”optimizer”,”nLogLik”,”n_parameters”,”aic”,tp,tx,”x”]
-
karenina.fit_timeseries.fit_input(input, ind, tp, tx, method)¶ Fit and minimize the timeseries for each subject defined
Parameters: - input – dataframe: [#SampleID,individual,timepoint,treatment,pc1,pc2,pc3]
- ind – subject column identifier
- tp – timepoint column identifier
- tx – treatment column identifier
- method – “basinhopping” if not defined in opts
Returns: dataframe: [ind,”pc”,”sigma”,”lambda”,”theta”,”optimizer”,”nLogLik”,”n_parameters”,”aic”,tp,tx,”x”]
-
karenina.fit_timeseries.fit_normal(data)¶ Return the mean and standard deviation of normal data
Parameters: data – fit data to normal distribution Returns: mu, std, nLogLik
-
karenina.fit_timeseries.fit_timeseries(fn_to_optimize, x0, xmin=array([-inf, -inf, -inf]), xmax=array([inf, inf, inf]), global_optimizer='basinhopping', local_optimizer='Nelder-Mead', stepsize=0.01, niter=200, verbose=False)¶ Minimize a function returning input & result fn_to_optimize – the function to minimize. Must return a single value or array x.
x0 – initial parameter value or array of parameter values xmax – max parameter values (use inf for infinite) xmin – min parameter values (use -inf for infinite) global_optimizer – the global optimization method (see scipy.optimize) local_optimizer – the local optimizer (must be supported by global method)
Parameters: - fn_to_optimize – function that is being optimized, generated from fn_to_optimize
- x0 – initial parameter value or array of parameter values
- xmin – min parameter values (-inf for infinite)
- xmax – max parameter values (inf for infinite)
- global_optimizer – global optimization method
- local_optimizer – local optimization method (must be supported by global)
- stepsize – size for each step (.01)
- niter – number of iterations (200)
- verbose – verbose output, default = False
Returns: global_min, f_at_global_min
-
karenina.fit_timeseries.gen_output(fit_ts, ind, tp, tx, method)¶ Generate output dataframe for either cohort, or non-cohort data
Parameters: - fit_ts – dataframe [0: [[Sigma, Lambda, Theta], nLogLik], 1: Individuals, 2: Times 3: Treatments, 4: Values, 5: PC Axis
- ind – individual identifier
- tp – timepoint identifier
- tx – treatment identifier
- method – optimization method
Returns: Formatted dataframe for csv output
-
karenina.fit_timeseries.get_OU_nLogLik(x, times, Sigma, Lambda, Theta)¶ Return the negative log likelihood for an OU model given data x – an array of x values, which MUST be ordered by time times – the time values at which x was taken. MUST match x in order.
OU model parameters
- Sigma – estimated Sigma for OU model (extent of change over time)
- Lambda – estimated Lambda for OU model (tendency to return to average position)
- Theta – estimated Theta for OU model (average or ‘home’ position)
Parameters: - x – array of values ordered by time
- times – times associated with x values
- Sigma – initial sigma value
- Lambda – initial lambda value
- Theta – initial theta value
Returns: nLogLik value
-
karenina.fit_timeseries.make_OU_objective_fn(x, times, verbose=False)¶ Make an objective function for use with basinhopping with data embedded
scipy.optimize.basinhopping needs a single array p, a function, and will minimize f(p). So we want to embded or dx data and time data in the function, and use the values of p to represent parameter values that could produce the data.
Parameters: - x – individual x values for objective function
- times – timepoints associated with passed-in x values
- verbose – verbose output, default = False
Returns: fn_to_optimize
-
karenina.fit_timeseries.make_OU_objective_fn_cohort(x, times, verbose=False)¶ Sums nLogLik and build objective function based on cohorts. Operates in the same manner as make_OU_objective_fn, except that it considers a treatment cohort, not just the individuals.
Make an objective function for use with basinhopping with data embedded
scipy.optimize.basinhopping needs a single array p, a function, and will minimize f(p). So we want to embded or dx data and time data in the function, and use the values of p to represent parameter values that could produce the data.
Overall strategy: Treat this exactly like the per-individual fitting, BUT within the objective function loop over all individuals in a given treatment (not just one) to get nLogLiklihoods. The nLogLiklihood for the individuals in the treatment is then just the sum of the individual nLogLikelihoods for each individuals timeseries.
data – a dict of {“individual1”: (fixed_x,fixed_times)}
Parameters: - x – x values for cohort objective function
- times – times associated with x values from cohort
- verbose – verbose output, default = False
Returns: fn_to_optimize
-
karenina.fit_timeseries.parse_metadata(metadata, individual, timepoint, treatment, site)¶ Parse relevant contents from metadata file to complete input dataframe
Parameters: - metadata – tsv file location
- individual – subject column identifier
- timepoint – timepoint column identifier
- treatment – treatment column identifier
- site – [subjectID, x1, x2, x3]
Returns: input dataframe: [#SampleID,individual,timepoint,treatment,pc1,pc2,pc3]
-
karenina.fit_timeseries.parse_pcoa(pcoa_qza, individual, timepoint, treatment, metadata)¶ Load data from PCoA output in Qiime2 Format
Parameters: - pcoa_qza – Location of PCoA file
- individual – Subject column identifier(s) [ex: Subject; Subject,BodySite]
- timepoint – Timepoint column identifier
- treatment – Treatment column identifier
- metadata – optionally defined metadata file location, if not defined, will use metadata from PCoA.qza
Returns: input dataframe, tsv filepath location
benchmark.py¶
-
karenina.benchmark.benchmark_simulated_dataset(n_timepoints, verbose=False, simulation_type='Ornstein-Uhlenbeck', simulation_params={'delta': 0.25, 'lambda': 0.12, 'mu': 0.5}, local_optimizer=['L-BFGS-B'], local_optimizer_niter=[5], local_optimizer_stepsize=0.005, log=[], dt=0.1)¶
-
karenina.benchmark.benchmark_simulated_datasets(max_timepoints=300, output_dir=None, simulation_type='Ornstein-Uhlenbeck', simulation_params={'delta': 0.25, 'lambda': 0.12, 'mu': 0.5}, local_optimizer=['L-BFGS-B'], niter=[5], verbose=False)¶ Test the ability of fit_timeseries to recover correct OU model params
Parameters: - output – location for output log
- max_timepoints – maximum timepoints to test. The benchmark will test every number of timepoints up to this number. So specifying max_timpoints = 50 will test all numbers of timepoints between 2 and 50.
- simulation_type – string describing the model for the simulation (passed to a Process object)
- simulation_params – dict of parameters for the simulation (these will be passed on to the Process object).
- local_optimizer – list of names of local optimizers to use
- niter – list of niter values to try. This parameter controls the number of independent restarts for global optimizer
- verbose – verbosity
Returns: output dataframe of benchmarked data
-
karenina.benchmark.calculate_errors(observed, expected, parameter_names=None)¶ Return per-parameter absolute and squared errors to results
Parameters: - names (parameter) – list of strings describing parameter names (in order)
- observed – an array of observed values
- expected – an array of expected values
-
karenina.benchmark.graph_absolute_errors(df, output_dir, model_name_col='model')¶ Graph absolute error in simulated datasets with varying numbers of tested timepoints
Parameters: - df – Dataframe containing model, timepoints, and list of errors
- output_dir – output directory
- model_name_col – column of the datafram that has the name of each model/benchmark
-
karenina.benchmark.log_lines_from_result_dict(results)¶ Return a list of result lines
Parameters: results – a dict of results (all values must be able to be converted to strings)
-
karenina.benchmark.make_scatterplot(x_col, y_col, data, hue, palette=['firebrick'], title='', outfile='./results_scatter.png')¶
-
karenina.benchmark.write_logfile(output_fp, log_lines)¶ Write logfile lines to output filepath
Parameters: - output_fp – a filepath (as string) for the output log file
- log_lines – lines of the logfile (linebreaks will be appended)
visualization.py¶
-
karenina.visualization.get_timeseries_data(individuals, axes=['x', 'y', 'z'])¶ Provides timeseries data from list of individuals.
Parameters: - individuals – array of individual objects
- axes – [x,y,z] axes to return
Returns: array of timeseries data
-
karenina.visualization.save_simulation_data(data, ids, output)¶ Saves simulation output data in PCoA format
Parameters: - data – data to save
- ids – Sample_IDs
- output – output filepath
-
karenina.visualization.save_simulation_figure(individuals, output_folder, n_individuals, n_timepoints, perturbation_timepoint)¶ Save a .pdf image of the simulated PCoA plot
Parameters: - individuals – array of individuals
- output_folder – output filepath
- n_individuals – number of individuals
- n_timepoints – number of timepoints
- perturbation_timepoint – timepoint of perturbation application
-
karenina.visualization.save_simulation_movie(individuals, output_folder, n_individuals, n_timepoints, black_background=True, data_o=False, verbose=False)¶ Save an .ffmpg move of the simulated community change
Parameters: - individuals – array of individuals to visualize
- output_folder – output directory filepath
- n_individuals – number of individuals
- n_timepoints – number of timepoints
- black_background – T/F, default = True
- verbose – verbose output, default = False
-
karenina.visualization.update_3d_plot(end_t, timeseries_data, ax, lines, points=None, start_t=0)¶ Updates visualization 3d plot
Parameters: - end_t – end timepoint
- timeseries_data – data from timeseries
- ax – visualization ax
- lines – lines of data
- points – values to update
- start_t – start timepoint (0)
experiment.py¶
-
class
karenina.experiment.Experiment(treatment_names, n_individuals, n_timepoints, individual_base_params, treatment_params, interindividual_variation, verbose)¶ Bases:
objectThis class has responsibility for simulating an experimental design for simulation.
A fixed number of individuals are simulated across experimental conditions called ‘treatments’ Each treatment can have different numbers of individuals. All treatment must have the same number of timepoints.
Each treatment can be associated with the imposition of one or more Perturbations. Each perturbation is inserted or removed from all individuals at a fixed time-point.
-
check_n_timepoints_is_int(n_timepoints)¶ Raise a ValueError if n_timepoints can’t be cast as an int
Parameters: n_timepoints – number of timepoints
-
check_variable_specified_per_treatment(v, verbose=False)¶ Raise a ValueError if v is not the same length as the number of treatments
Parameters: - v – variable
- verbose – verbose output, default = False
-
q2_data()¶ generate output data from object’s self for Qiime2
Returns: data, ids
-
run()¶ Run the experiment, simulating timesteps; saves the simulation figure and movie
-
simulate_timestep(t)¶ Simulate timestep t of the experiemnt
Approach:
- First apply any perturbations that should be active but aren’t yet
- Second, simulate the timestep
- Third, remove any perturbations that should be off
NOTE: this means that a perturbation that starts and ends at t=1 will be active at t=1 That is, the start and end times are inclusive.
Parameters: t – timestep
-
simulate_timesteps(t_start, t_end, verbose=False)¶ Simulate multiple timesteps
Parameters: - t_start – start timepoint
- t_end – end timepoint
- verbose – verbose output, default = False
-
write_to_movie_file(output_folder, verbose=False)¶ Write an MPG movie to output folder
Parameters: - output_folder – output directory
- verbose – verbose output, default = False
-
individual.py¶
-
class
karenina.individual.Individual(subject_id, coords=['x', 'y', 'z'], metadata={}, params={}, interindividual_variation=0.01, verbose=False)¶ Bases:
objectGenerates an individual for OU simulation
-
apply_perturbation(perturbation)¶ Apply a perturbation to the appropriate axes
Parameters: perturbation – perturbation to apply
-
apply_perturbation_to_axis(axis, perturbation)¶ Apply a perturbation to a Processes objects
Parameters: - axis – Axis to apply perturbation to
- perturbation – perturbation to apply
-
check_identity(verbose=False)¶ Check identity of movement process
Parameters: verbose – verbose output, default = False Returns: True if processes are equivalent, False if not
-
get_data(n_timepoints)¶ get data from movement processes
Parameters: n_timepoints – number of timepoints to gather data for Returns: data for timepoints
-
remove_perturbation(perturbation)¶ Remove a perturbation from the appropriate axes
Parameters: perturbation – perturbation to remove
-
remove_perturbation_from_axis(axis, perturbation)¶ Remove a perturbation from one or more Process objects
Parameters: - axis – axis to remove perturbation from
- perturbation – perturbation to remove from axis
-
simulate_movement(n_timepoints, params=None)¶ Simulate movement over timepoints
Parameters: - n_timepoints – number of timepoints to simulate
- params – parameters to change from baseparams
-
-
karenina.individual.random() → x in the interval [0, 1).¶
perturbation.py¶
-
class
karenina.perturbation.Perturbation(start, end, params, update_mode='replace', axes=['x', 'y', 'z'])¶ Bases:
objectAlter a simulation to impose a press disturbance, shifting the microbiome torwards a new configuration
start –inclusive timepoint to start perturbation. Note that this is read at the Experiment level, not by underlying Process objects.
end – inclusive timepoint to end perturbation. Note that this is read at the Experiment level, not by underlying Process objects.
params – dict of parameter values altered by the disturbance
- ‘mode’ – how the perturbation updates parameter values.
- ‘replace’ – replace old value with new one
- ‘add’ – add new value to old one
- ‘multiply’ – multiply the two values
axes – axes to which the perturbation applies. Like Start and End this is a ‘dumb’ value, read externally by the Experiment object
-
is_active(t)¶ determines if timepoint is active
Parameters: t – timepoint Returns: True if timepoint is active, False if not
-
update_by_addition(curr_param, perturbation_param)¶ Update parameters by addition
Parameters: - curr_param – current parameter
- perturbation_param – new parameter
Returns: curr_param + perturbation_param
-
update_by_multiplication(curr_param, perturbation_param)¶ Update parameters by multiplication
Parameters: - curr_param – current parameter
- perturbation_param – new parameter
Returns: curr_param * perturbation_param
-
update_by_replacement(curr_param, perturbation_param)¶ Update parameters by replacement
Parameters: - curr_param – current parameter
- perturbation_param – new parameter
Returns: perturbation_param
-
update_params(params)¶ Update baseparams
Parameters: params – params to update Returns: new_params
process.py¶
-
class
karenina.process.Process(start_coord, motion='Ornstein-Uhlenbeck', history=None, params={'L': 0.2, 'delta': 0.25})¶ Bases:
objectRepresents a 1d process in a Euclidean space
-
bm_change(dt, delta)¶ Change the Brownian motion process
Parameters: - dt – time elapsed since last update
- delta – delta value to adjust
Returns: change
-
bm_update(dt, delta)¶ Update the Brownian motion process
Parameters: - dt – time elapsed since last update
- delta – delta value to adjust
Returns: change
-
ou_change(dt, mu, L, delta)¶ Change the Ornstein Uhlenbeck motion process
The Ornstein Uhlenbeck process is modelled as:
ds = lambda * (mu - s) * dt + dW
ds – change in our process from the last timepoint L – lambda, the speed of reversion to mean (NOTE: lambda is a reserved keyword in Python so I use L) mu – mean position s – current position dt – how much time has elapsed since last update dW – the Weiner Process (basic Brownian motion)
This says we update as usual for Brownian motion, but add in a term that reverts us to some mean position (mu) over time (dt) at some speed (lambda)
Parameters: - dt – time elapsed since last update
- delta – delta value to adjust
Returns: change in process since last timepoint
-
ou_update(dt, mu, L, delta, min_bound=-1.0, max_bound=1.0)¶ Update the Brownian motion process
Parameters: - dt – time elapsed since last update
- mu – mean position
- L – speed of reversion to the mean
- delta – variance from the mean
- min_bound – minimum bound
- max_bound – maximum bound
-
update(dt)¶ Update the process
Parameters: dt – time elapsed since last update
-